Hello everyone,
I figured I should give an elaborate background of why I am using NDVI and how I am processing my images before I get to my questions, so here goes. If you are not interested in all this detail, please skip ahead to the bulleted list near the end subtitled 'My questions'.
An introduction about my project before I ask a few questions:
My research team and I are developing an inexpensive sensor that will automatically detect changes in phenotypes of plants due to environmental stressors. We are doing so with the use of a Raspberry Pi Zero W, along with the Raspberry Pi Camera V2 and the Raspberry Pi Camera NoIR V2. One of our primary goals is that our product should be easy to use and setup for all kinds of people and professions. Furthermore, it is important that it the image acquisition and analysis processes are automated so that, for example, if a fungal infection were to occur in a plant, it would alert the owner of this risk in order for them to prevent an outbreak.
My goals for NDVI:
Because we require a large degree of automation, we need to write a workflow that will be able to tell which objects in an image are the plants that we wish to analyze, and which are not. This should be done without the need for any user input. After some research online, it seems like NDVI is the perfect tool to do so! So, I need my NDVI images to accurately pick out plant objects and nothing else. Although, if it keeps a few non-plant pixels, I will not be too concerned.
My setup:
I currently have my VIS camera outdoors (in a waterproof box) pointing at a mint plant. My NIR camera is pointing at the same plant, but at a 90˚ angle to the VIS camera. The NIR camera has a blue filter on the outside of it, which came with the camera. I believe this is the Rosco #2007 filter, but I could be wrong. Currently, I am creating NDVI images using the one NIR camera by treating the 'R' channel as NIR light, and the 'B' channel as VIS light.
My process:
I tried manually changing the white balance values of my camera, but this best value I came up with seems to be about (1.18, 0.74). I am not changing the exposure or ISO. Here is an image taken with these settings. (You can also see another bush in the background)
 ****____
****____
****____
I am doing the image processing in Python. Here is how my workflow goes:
- I iterate over each pixel in the image. Each pixel gets assigned an NDVI value equal to (R-B)/(R+B)
- The NDVI values are on the interval [-1, 1], so I map them to the interval [0, 255] so each pixel gets a grayscale value, which I make an image out of.
- Then, I am using the cfastie LUT to map these grayscale values to (R, G, B) values to get an NDVI image with colour.
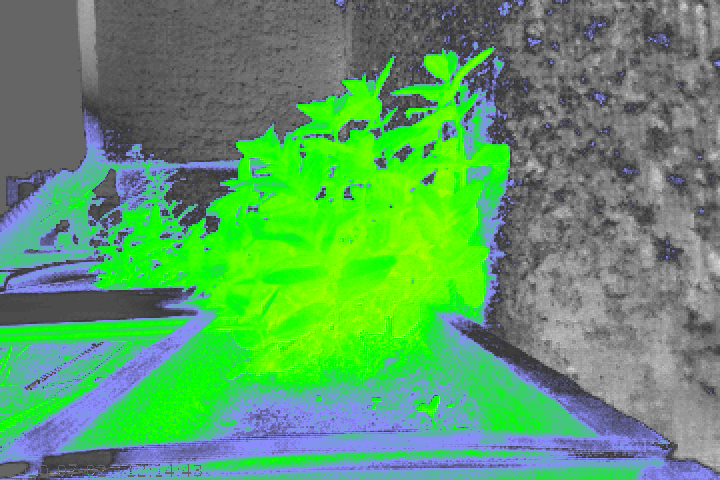
Here are the grayscale and coloured NDVI images corresponding to the above image.
 ****____
****____
 ****____
****____
****____
**
The problems:
**
As you can see, there are many objects in the image that are being assigned vegetation NDVI values which do not actually have photosynthesis activity. Additionally, the mint plant itself does not seem to have as high an NDVI value as I would expect. For example, I imagine it should probably be yellow for the most part instead of green.
Evidently this workflow is functioning to some degree, as the plant is clearly highlighted, but as stated above, I need a higher degree of accuracy in order to use this function long-term.
**
My questions:
Are other non-plant objects likely being assigned higher NDVI values due to NIR light bleeding into the 'B' channel?
Would a red filter instead of a blue one largely fix this problem?
As you can see in the grayscale, there really is not much contrast in NDVI values overall. They all seem to be within a close range to 0. Why is this the case, and how could I improve this?
If I were instead to try creating an NDVI image using a 2 camera setup, where 1 camera is VIS and 1 is NIR, which channels do I use to compute the NDVI value of each pixel? For example, should I simply use the 'R' channel on the VIS camera, and the 'R' (i.e. NIR) channel on the NIR camera with the same blue filter?
Would having targets with known reflectance help to eliminate this problem? Or would it not be enough on its own?
Thank you for reading through my whole post and I thank you all in advance for your input. I am very excited about getting NDVI working as I think it is an incredibly powerful tool that has yet to see its full potential in automated field phenomics.
Hey @velhas! Your work is super fascinating and useful! I come from a GIS background so I'm not too familiar with using NDVI in the way that you are presenting in this post, but thought I should suggest looking into an unsupervised classification program using python to automate identification of vegetation and masking of background pixels. This way you can identify a threshold for infrared reflectance values and have those classified as vegetation while other pixels can be classified otherwise.
I couldn't find an exact match when I searched for someone doing this already, but I found this resource that may be helpful in your coding: https://www.spectralpython.net/algorithms.html
I've only used aerial nadir satellite or UAS collected NDVI data, so I'm not sure if things are 1:1 with what you're doing, but I've always encountered NDVI data is usually clustered around 0 with few outliers spanning to the -1 and 1 values. In GIS you can manipulate the color ramps and the max / min values to emphasize variation in NDVI value, usually my threshold would be something like -.5 - +.5, even as precise as -.1 - + .1 for one project. Since reflectance is a remotely sensed value I always assumed the lack of spread was due to distance away from the plant reflecting IR light, but that's just my own conjecture.
I hope this helps! I've learned most of what I know about remote sensing through my time with NASA DEVELOP, if you want another resource of experts doing experimental multispectral work + thorough documentation!
Hi @a1ahna,
Thank you for your suggestions! Since reading your post, I've begun to look into Spectral Python and it seems like it could be of great value. Seeing as there is so much documentation for me to go through, I was wondering if you might be able to lead me towards which functions in SPy might be of the most importance for my purposes.
I am glad to hear that I am not the only one with NDVI values hovering around 0 for all my images! Is emphasizing variation in NDVI values something that SPy can do? Or did you use other resources to do so? Since my post, I've begun normalizing my grayscale NDVI images, and then producing a colour map with the cfastie LUT. This exaggerates the NDVI values, but perhaps your method may work better.
For example, here is the normalized version of the grayscale image in my original post:
And here is the NDVI image with a colour map that the normalized grayscale produces: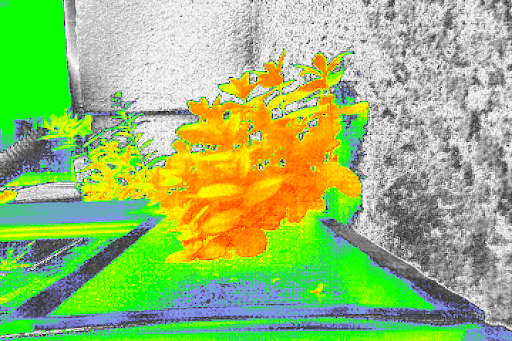
Is this a question? Click here to post it to the Questions page.
Hi @velahs,
The improvements you have obtained are really good.
I would like to know, what is the process that you have followed to improve your first NDVI image, and generate the second one in which the plants show yellow and orange colors? In other words, how did you normalize the gray scales to produce the last image of the NDVI values?
Is this a question? Click here to post it to the Questions page.
Reply to this comment...
Log in to comment
@aussie11950 awards a barnstar to velahs for their awesome contribution!
Reply to this comment...
Log in to comment